WHAT'S NEW?
2021-02-05
FutureFlu is recommending the following vaccines for the 2021-22 flu season. A/Virginia/11681/2020 and A/Macao/600028/2020 for the H1N1pdm and H3N2 components of the vaccine, respectively.
2020-11-20
FutureFlu is augmenting its current biostatistical model of the influenza virus. The new model is expected to be more precise in tracking and predicting the future flu strains. It will be used for all analyses hereafter. The recommended vaccine for the 2021-22 flu season, based on the new model, will be announced here in January 2021. WHO/CDC customarily declares their prescription for the upcoming flu season in the February-March timeframe. Based on the analysis till now, FutureFlu expects the vaccine to be in the clades 6A.1 and 3C.3a for the H1N1pdm and H3N2 components of the vaccine, respectively.
2020-10-11
The post hoc analysis shows that the A/Kansas/14/2017 subclade (3C.3a) was a good choice by WHO/CDC for the H3N2 component of the 2019-20 flu vaccine . While both clades 3C.3a and 3C.2a were prominent in the samples collected in 2020, the former seems to be taking a lead. Among the samples collected after 2020-01-31, 58.1% belonged to clade 3C.3a while 42.0% were in clade 3C.2a. Further breakdown places 18.5% in subclade 3C.2a1b+T131K, 17.5% in subclade 3C.2a1b+T135K-A and 6% in subclade 3C.2a1b+T135K-B.
2020-09-01
While developing new vaccines against the influenza virus remains an illusive task, the neuraminidase inhibitors (NI) continue to be effective. NI prevents a mature virus from leaving the infected cell and spreading the infection in a person. The infected cells are eventually killed by the host immune system along with their load of viruses. The popular NIs include oseltamivir (tradename Tamiflu) and zanamivir (tradename Relenza). In the 2,292 samples, NIs were >99% effective. The samples constituted 942 A/H1N1pdm, 794 A/H3N2 and 556 Type B viruses. It implies that unlike the vaccines, a single NI is viable across all major types of flu.
2020-07-14
On the H1N1pdm side, the clade 6B.1A5 is continuing its bifurcation into subclades 6B.1A5A and 6B.1A5B. 6B.1A5A is defined by N260D with additional mutations N129D and T185I in HA1. On the other hand 6B.1A5AB is identified by E235D and N260D with K130N, K160M, T216K and H296N in HA1.
2020-05-27
Even though the 2019-20 flu season has wound down, the labs around the world are still processing the samples collected in 2020. WHO/CDC had changed the H3N2 component of the 2019-20 vaccine from the A/Singapore/INFIMH-16-0019/2016 subclade (3C.2a1) to A/Kansas/14/2017 subclade (3C.3a). According to the analysis of the reported data, the two clades are co-dominating at this point. Among 780 samples of H3N2 analyzed, 52.85% and 47.17% belonged to clades 3C.3a and 3C.2a1b, respectively.
2020-03-11
FutureFlu's earlier argument that A/Brisbane/02/2018 was not an appropriate vaccine for 2019-20 was reinforced by the data collected between weeks 40/2019 to 06/2020. Among the 656 samples of H1N1 viruses that were characterized, 600 (91.46%) fell in the A/Norway/3433/2018 subclade (6B.1A5A). Only 8 (1.21%) belonged to the A/Brisbane/02/2018 subclade (6B.1A1).
2020-01-03
FutureFlu is augmenting its biostatistical model of the influenza virus with artificial intelligence. Therefore, there will be no recommendation of flu vaccine for the 2020-21 flu season. The recommendation the 2021-22 season will be announced in January 2021.
2019-10-29
A total of 1,885 H1N1 samples were collected and genetically characterized between weeks 40/2018 and 20/2019. All but three of the 1,885 samples fell in the A/Michigan/45/2015 vaccine component. This demonstrates that the H1N1 vaccine recommended by FutureFlu was closer to the prevalent strains than the one done by WHO/CDC. WHO/CDC had recommended A/Brisbane/02/2018.
2019-07-23
The mid year trend shows that the H1N1 strains are expected to continue drifting towards the genetically diverse phylogenetic clade 6B.1A. They carry the S183P substitution in HA1. On the H3N2 side, subclade 3C.3a is expected to dominate over the currently prevalent subclade 3C.2a1b .
2019-03-21
WHO/CDC announced the 2019-20 seasonal vaccines today. They recommended A/Kansas/14/2017 and A/Brisbane/02/2018 for H3N2 and H1N1, respectively. FutureFlu had previously calculated the vaccine as A/Netherlands/10160/2018 (H3N2) and A/Michigan/45/2015 (H1N1). FutureFlu's calculations were remarkably close to that of WHO/CDC. There was a 100% query coverage between WHO/CDC and FutureFlu for both vaccines. Correspondingly, there were 98.23% and 98.59% identities.
2018-12-12
FutureFlu recommends an A/Netherlands/10160/2018-like (NL18) strain for the H3N2 component of the 2019-20 flu vaccine. Further, A/Michigan/45/2015 (MI15), the current H1N1pdm vaccine should be continued. The FutureFlu model suggests that the new H3N2 vaccine should incorporate the N112S, K137N, T147K, G158K, K187N, R277Q, V422I, E495G and E500G mutations over the current vaccine.
2018-03-01
WHO/CDC announced their recommended vaccines for the 2018-19 flu season. Once again, the vaccines computed by FutureFlu were a perfect match with that of WHO/CDC. The BLASTP comparison of H3N2 strains A/Slovenia/342/2017 (recommended by FutureFlu) and A/Singapore/INFIMH-16-0019/2016 (recommended by WHO/CDC) shows 100% identities with 0% gaps.
2018-02-18
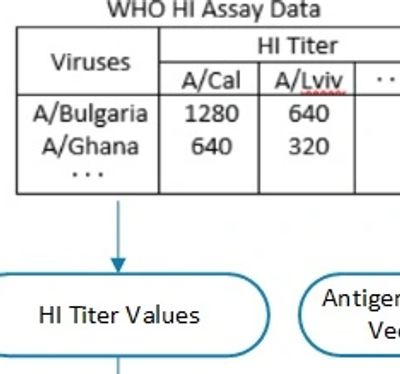
According to The Francis Crick Institute report, “H3N2 viruses have become increasingly difficult to characterize antigenically by HI assay”. HI (hemagglutinin inhibition) assay relies on the agglutination (i.e. clumping) of red blood cells (RBCs) by the influenza virus. It is used for calculating the efficacy of an influenza vaccine against a virus strain.
The report states that the HA assay is losing viability because H3N2 viruses are no longer agglutinating the RBCs effectively. Also, the results are becoming less repeatable.
Above findings support the need for supplementing the HI assay used by WHO with alternate methods such as the reverse vaccinology model developed by FutureFlu.
2018-01-02
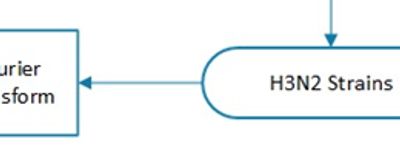
FutureFlu recommends an A/Slovenia/342/2017-like (SL17) strain for the H3N2 component of the 2018-19 flu vaccine. Further, A/Michigan/45/2015 (MI15), the current H1N1pdm vaccine should be continued. The composition of the H3N2 vaccine was calculated with the Fourier-enhanced Phylogeny/Antigenicity Predictive Model (F^P/A-PM) developed by FutureFlu. The model suggests that the new vaccine should incorporate the N120K, R141G, N170K, I405V, G478E and G483E mutations over the current vaccine.
2017-09-05
According to the WHO Weekly Report dated 04 Sep 2017, Type A influenza (H1N1pdm and H3N2) continues to dominate the flu sickness. 89.2% of the reported cases are affected by it. FutureFlu is extending the Prediction Model to H3N2, with an emphasis on epitopes A-E on the HA segment of H3.
2017-04-20
FutureFlu had recommended A/Michigan/45/2015 as the H1N1pdm component of the flu vaccine in January 2017. It belongs to the Clade 6B.1. How does it compare to the emerging Clade 6B.2? The analysis using the Antigenic Prediction Model shows that the drift between the two Clades is still within the limits of vaccine efficacy.
2017-03-07
The World Health Organization has announced the vaccine for the 2017-18 flu season. The A/Michigan/45/2015 strain recommended by WHO is a perfect match with A/Montana/04/2016 recommended by FutureFlu.
2017-03-01
Strains from Clade 6B.1 continue to dominate H1N1pdm (www.cdc.gov/flu/weekly/pdf/External_F1708.pdf). A/Montana/04/2016 is much more closely associated with Clade 6B.1 genetically than the current vaccine strain A/California/07/2009 .
2017-02-12
In their Weekly U.S. Influenza Surveillance Report dated February 10, 2017, CDC stated “The HA gene segment of all influenza A (H1N1)pdm09 viruses analyzed belonged to genetic group 6B.1” (www.cdc.gov/flu/weekly/pdf/External_F1705.pdf).
The A/Montana/04/2016 vaccine recommended by FutureFlu belongs to the same Clade 6B.1. It further supports the analysis by FutureFlu and the need for a new vaccine.
2016-12-15
Based on the phylogenetic and antigenicity analyses, FutureFlu recommends an A/Montana/04/2016-like vaccine for the 2017-18 flu season, preferably using a recombinant specimen.
2016-09-12
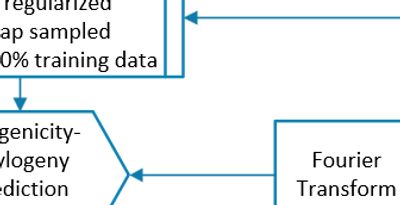
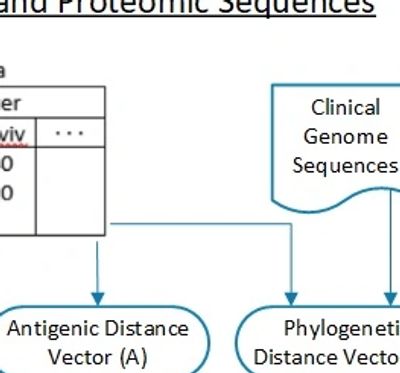
FutureFlu has developed a novel Antigenicity Prediction Model of the hemagglutinin inhibition (HI) assay. It estimates the avidity of the antibodies generated by a vaccine towards an influenza strain. It uses the proteomic sequences of the strains and the antisera.
© 2021 stemXchange LLC - All Rights Reserved.